Peptide Mapping
Complete Sequence Coverage and High Throughput Analysis of Multiple CQAs
What is Peptide Mapping?
Peptide mapping is a key application for characterization of biotherapeutic proteins such as monoclonal antibodies (mAbs), antibody-drug conjugates (ADCs), and viral vector capsids in drug discovery and process development.
The purpose of peptide mapping is for confirmation of protein’s primary structure, identify and monitoring post translational modifications (PTMs), and for assessing biosimilarity to innovator drug product. Robust peptide mapping workflow is critical to ensuring quality and reproducibility of biotherapeutic proteins with fast turnaround time being highly desirable.
ZipChip is an out of the Box Solution for Fast and Easy Peptide Mapping Including Multi-Attribute-Monitoring(MAM):
- Near 100% sequence coverage and high-res peptide separation within 15 minutes
- Easily resolve glycopeptides, deamidation, oxidation, and other site-specific PTMs
- Enhanced throughput for MAM (up to 20x)
- No method development with pre-optimized ready to use essay kits
Higher Sequence Coverage for Higher Confidence Data
Peptide Mapping byZipChip can easily determine sequence changes and get close to 100% sequence coverage of both light and heavy polypeptide chains and catch hydrophilic peptides, which often are not retained by reversed phase LC (RPLC).
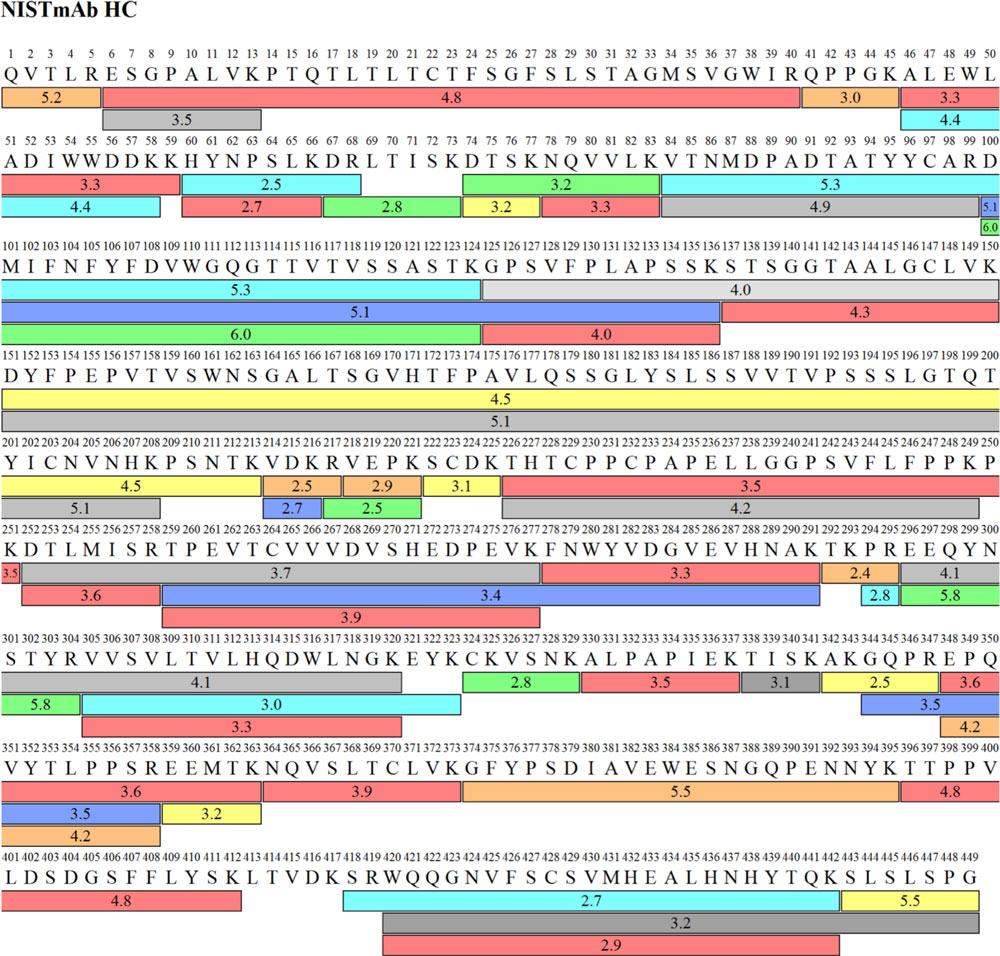
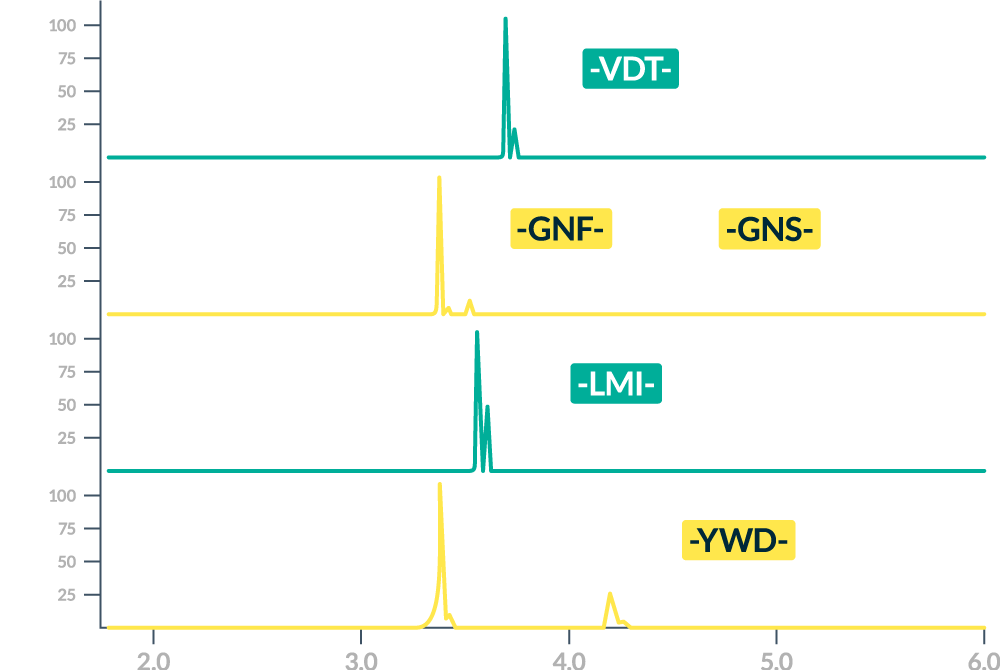
Quickly Resolve PTMs and Identify Key CQAs
ZipChip-MS is an ideal technique for separating tricky analytes like conjugated peptides, complex glycopeptides, isomerized residues, and degradants.
Quickly resolve and get actionable information on critical quality attributes (CQAs) such as glycopeptides, deamidation, oxidation and other site-specific modifications in less than 15 minutes. With an ZipChip autosampler (Optional), simply load the samples, press go and come back later for results.
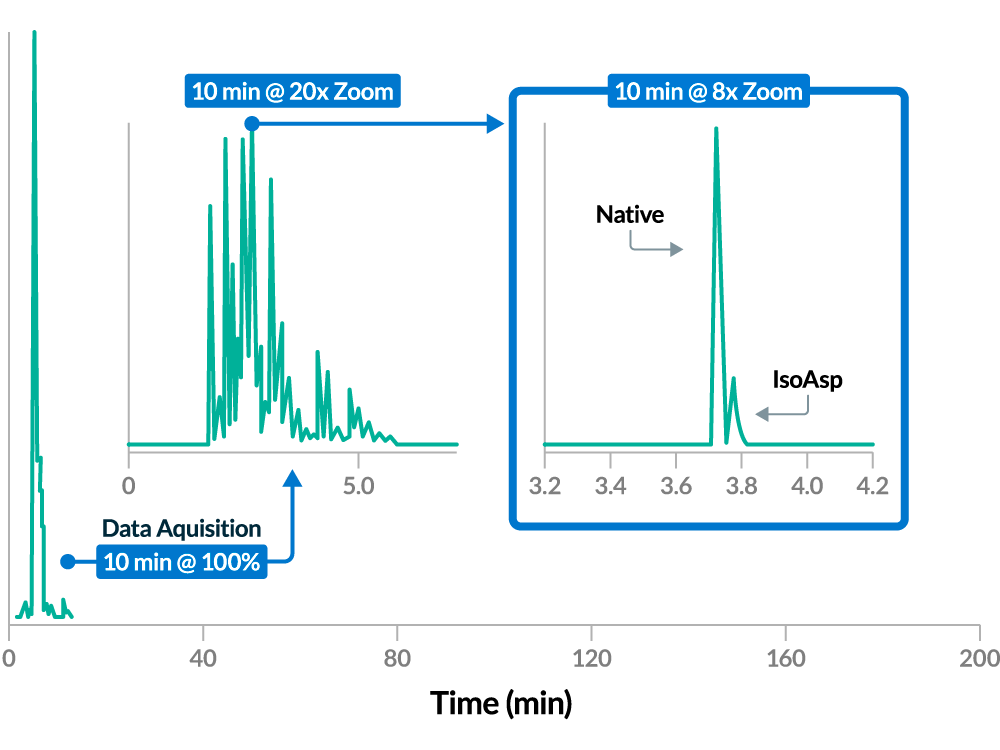
Enhanced Throughput of MAM
Compared with LC-MS, ZipChip CE-MS can be up to 20x faster, with higher sequence coverage, and comparable quantitation for a panel of 15 CQAs including deamidation, aspartic acid isomerization, methionine oxidation, and tryptophan oxidation.
1.Chip-Based Capillary Zone Electrophoresis Mass Spectrometry for Rapid Resolution and Quantitation of Critical Quality Attributes in Protein Biotherapeutics. Andrew B. Dykstra, Et al. Journal of the American Society for Mass Spectrometry 2021.
No Method Development
Pre-optimized ready to use essay kits practically eliminate the need for method development. Detergents and salts in protein sample matrix wreak havoc on LC-MS systems, not on ZipChip though. The ZipChip assay performance is not affected by detergents and salts from matrix and there is no risk of sample carryover.

Get Straight to it.
ZipChip consumables take the guesswork out of method development. Assay kits come with background electrolyte and sample diluent that’s optimized for each application. Just put the bottle of background electrolyte in the autosampler, prep your samples with the sample diluent and you’re off!
How to Find Your Assay & Consumables Kit:
Pick a BGE Kit
Then a Chip
Super-charge Your Mass Spec
Struggling with increased workloads? Increase your throughput with plug and play applications for multiple Critical Quality Attributes (CQAs). Maximize your MS investment and boost your lab’s productivity with multiple characterization assays. Whether you’re running intact charge variant analysis, subunit analysis, peptide mapping, metabolites analysis, or oligonucleotides analysis, ZipChip works at every level.
CE Mass Spectrometry
ZipChip
Pop ZipChip on your mass spec as the source. It separates your samples and electrosprays them into your mass spec for analysis. There’s no optimization to do or connections to make. Click the chip in place and you’re off. Ready to go assay kits are optimized for each application, all you do is dilute-and-shoot. From peptide mapping to native charge variants analysis, ZipChip can do it all. And you don’t have to spend extra time for method development.

Suggested Resources

Microchip CE/MS Peptide Mapping Workflow with Trypsin Digestion for the Analysis of Critical Quality Attributes of Monoclonal Antibodies
Peptide mapping is considered the gold standard for the analysis of biopharmaceuticals, including monoclonal antibodies.

Application Experience: Native Analysis of mAbs and ADCs

Customer Experience: New England Biolabs

Fast Therapeutic Protein Characterization
Frequently Asked Questions
What sample preparation is needed to run Peptide Mapping on ZipChip?
In order to break a protein down into peptides, there are a few steps that need to be followed, such as denaturation, reduction, alkylation, and enzymatic digestion. This is still true for performing Peptide Mapping on the ZipChip, however, we have developed techniques to make this process as straight-forward and rapid as possible. Our Peptide Mapping protocol includes detailed steps that allows for a sample to be ready for analysis within two hours as opposed to the several days most protein digestion protocols call for.
What method development would I need to do to run Peptide Mapping on ZipChip?
Our Peptide Mapping specific protocol includes guidelines for sample preparation, system suitability testing, mass spectrometer method parameters, and ZipChip separation method parameters. These guidelines tend to generate robust and accurate data, but every mass spectrometer is different and there might be some slight modifications that need to take place for optimal data collection. This is usually very minimal, even with more complex samples such as a protein digest.
What is the run time for Peptide Mapping on ZipChip?
A run time of 10-15 minutes is very common for running a Peptide Mapping analysis of a protein or mAb using ZipChip. Compare this to one or more hours on an LC-MS setup, and the speed benefit becomes very apparent.
What Critical Quality Attributes am I able to characterize when running Peptide Mapping on ZipChip?
Digesting a protein into smaller peptides allows for more detailed post-translational modifications to be recorded. PTMs, such as deamidation, Met Oxidation, Trp Oxidation, Isomerization of Aspartic Acid, Glycosylation, Succinimide Intermediates, or Dioxidation, are all PTMs that have been recorded using the ZipChip coupled to a high resolution mass spectrometer.
What sequence coverage can I expect of my protein when running Peptide Mapping on ZipChip?
Usual sequence coverage for a digested protein or monoclonal antibody (mAb) when separated on a ZipChip is between 95-100%. In contrast, the sequence coverage using an LC-MS system is usually in the low 90%. The reason is that smaller peptides (such as di or tri peptides) tend to not be retained on the stationary phase of an LC, thus not being properly separated and recorded in the mass spectrometer. Since the ZipChip microfluidic chip utilizes an open capillary to separate analytes by charge and size, this is not an issue, and small di and tri peptides do show up in the data, resulting in generally higher sequence coverage of a protein compared to LC-MS.
Is quantitation of peptides possible when using ZipChip for separation?
Yes! The ZipChip utilizes your mass spectrometer’s data collection software, so the output format would be similar to any other separation done using that mass spectrometer as the analyzer. Data analysis software such as Skyline, Proteome Discoverer, MaxQuant, Mascot, MultiQuant, Analyst, Proteoscape, Biopharma Finder, Biopharma ComPass, or any other software capable of quantifying peptides, can be used to process data generated by ZipChip coupled to a mass spectrometer.
What are the benefits of running Peptide Mapping on ZipChip over more traditional methods such as LC-MS?
There are two major benefits of performing Peptide Mapping on the ZipChip over running it on a Liquid Chromatography – Mass Spectrometry setup. First, the ZipChip separation is extremely fast. A Peptide Mapping analysis that can take hours on an LC-MS system takes minutes on the ZipChip. Second, smaller peptides (such as di or tri peptides) tend to not be retained on the stationary phase of an LC, thus not being properly separated and recorded in the mass spectrometer. Since the ZipChip microfluidic chip utilizes an open capillary to separate analytes by charge and size, this is not an issue, and small di and tri peptides do show up in the data, resulting in generally higher sequence coverage of a protein compared to LC-MS.
email Subscribe to Our Communications Signup to receive new product updates, technical tips and more.